HOW CAN WE HELP YOU? Call 1-800-TRY-CHOP
In This Section
A ‘Trojan Horse’ to the Rescue of Staph Infections
Using structure-guided design, Dr. Audrey Odom John and her research team developed a new way to design better antibiotics that are able to reach resistant bacteria.
Taking a cue from Homer’s Odyssey and the infamous story of the Trojan horse, researchers in the John Laboratory at Children’s Hospital of Philadelphia established a novel approach for developing effective antibiotics that are able to reach resistant bacteria. The study proposes structure-guided design to create prodrugs that conceal the antibiotics and then allow them to be revealed and released — by the bacteria themselves — once the antibiotics arrive at the site of infection. A Trojan horse, indeed. eLife recently published the study findings.
What makes this research novel is the way the investigators manage to target the bacteria’s own machinery. By investigating and understanding the mechanism by which the bacteria removes the blocking molecule, or prodrug, the CHOP researchers were able to design a better method of delivering the effective compound. This structure-guided drug design allows the antibiotic molecule — with the prodrug — to circulate in the bloodstream until reaching the bacteria where the blocking agent then comes off. The researchers believe they can improve the qualities of the antibiotic molecule without releasing the prodrug early and missing the intended target, which has been a major challenge in the effort to develop effective antibiotics.
“We’ve created a sort of ‘Trojan horse’ that would allow antibiotics to reach desired tissues undisturbed, until the bacteria itself activates the drug, effectively releasing an ‘army’ of antibiotics,” said senior author Audrey Odom John, MD, PhD, chief of the Division of Infectious Diseases. “Using structure-guided design, we have developed a new way to design better antibiotics. Given the growing concern over antimicrobial resistance, we think this is an important step forward.”
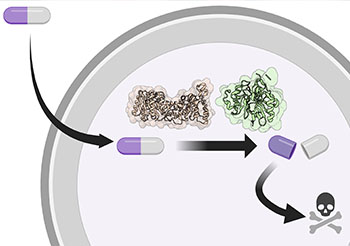
Delivering effective antibiotics to hard-to-reach resistant bacteria.
A Threat to Public Health
Antimicrobial resistance — when microorganisms such as bacteria, viruses, fungi, and parasites develop the ability to defeat the drugs designed to kill them, is one of the world’s most critical public health problems. Approximately 2.8 million people are infected with antimicrobial resistant germs in this country each year; more than 35,000 people die as a result, according to the U.S. Centers for Disease Control and Prevention. Worldwide, resistant bacteria cause over 750,000 deaths annually, with predictions for these numbers to increase into the millions.
There’s an urgent need for scientists to develop new antibiotics that can circumvent antimicrobial resistance. Unfortunately, attempts to develop new treatments often fail in animal models or clinical trials due to insufficient levels of treatment reaching the desired tissues.
In this study, Dr. John’s team concentrated on the bacteria Staphylococcus aureus, because methicillin-resistant S. aureus (MRSA) is labeled a “serious threat” by the CDC. In an attempt to understand just how staph bacteria remove prodrugs, they engineered the bacteria to do just the opposite — become resistant and not remove the prodrug. Then, by sequencing their entire genomes, the investigators were able to identify what genes were mutated in the bacteria that had become resistant. This knowledge led them to two different proteins (GloB and FrmB) that sequentially act to take the prodrug off — the new bacterial targets were found.
“This work paves the way for structure-guided development of S. aureus-specific prodrugs and establishes a pipeline for the identification of additional microbial prodrug activating enzymes,” said Dr. John, who is also on facutly in the Perelman School of Medicine at the University of Pennsylvania. “We anticipate that these approaches will both guide the development of novel antimicrobials and lead to a more robust arsenal of anti-infective compounds with targeted specificity for the microbe over the human host.”