HOW CAN WE HELP YOU? Call 1-800-TRY-CHOP
Campbell Laboratory Research Overview
The Campbell Lab uses electronic health record data to gain better understanding of the full scope of the features of genetic diseases, as well as to understand and ameliorate inequities in genetic testing and care. Some of the lab's current research projects are highlighted below.
Many genetic studies are performed using phenotypic information manually abstracted by clinicians or researchers. In some cases, phenotypes relevant to genetic studies are automatically extracted from electronic health record (EHR) billing codes. Few studies computationally extract information from free-form clinical text such as progress notes or imaging interpretations.
The Campbell Lab uses state-of-the-art machine learning techniques including pre-trained language models and recurrent neural networks to extract standardized genetic phenotypes from EHR data. We have enlisted multiple clinical specialists to produce annotated training data to train our models.
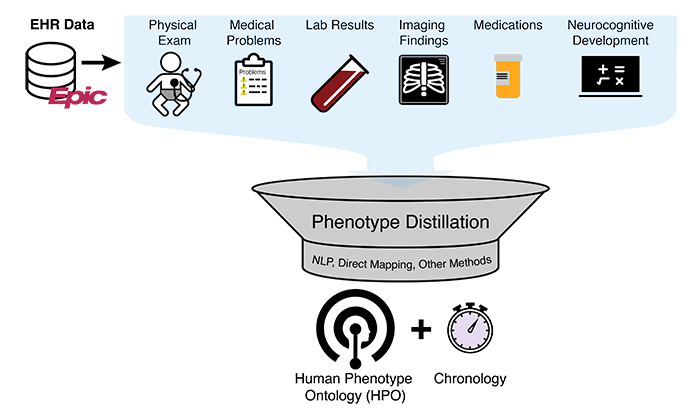
Clinicians have developed clinical diagnostic criteria that differentiate among 7,000 rare genetic diseases and enumerate different elements—medical problems, physical exam findings, laboratory test results, and imaging findings—that define the disease when appearing in combination.
For most diseases, existing diagnostic criteria have unknown predictive value. Despite being critical for diagnosis and genetic testing, they are typically proposed without rigorous evidence or estimates of performance. If criteria have suboptimal sensitivity, clinicians may overlook or dismiss diagnoses, depriving patients and families of prognostication, reproductive planning, or targeted therapeutic intervention. If criteria have suboptimal specificity, clinicians may arrive at incorrect diagnoses or make faulty interpretations of genetic testing with variants of uncertain clinical significance. We previously established a generalizable approach to developing evidence-based rare disease criteria using statistical optimization.
As proof of principle, we developed novel criteria for the hereditary cancer disease nevoid basal cell carcinoma syndrome (NBCCS) using survey data, assistance from a Bernoulli naive Bayes classifier, and clinical insight. These criteria have improved sensitivity compared to previous expert consensus criteria, particularly at early ages (53% vs. 13% at 7 years), while maintaining diagnostic specificity.
We hypothesize that diagnosis of rare pediatric genetic disease can be improved by implementing evidence-based diagnostic approaches. Thus, one of our principal goals is the implementation of statistical and machine learning frameworks to define diagnostic criteria and test their performance.
Gold NB, Campbell IM, Sheppard SE, Tan WH. Proposed criteria for nevoid basal cell carcinoma syndrome in children assessed using statistical optimization. Sci Rept. 2021 Oct 5;11(1):19791. PMID: 34611197
Making systematic improvements to the way that genetic care is delivered and reducing inequities is a major goal of the laboratory.
Through an analysis of patient characteristics to assess why some genetic test results had not been reviewed after completion, we found that patient-reported race was a significant predictor of documented genetic testing follow up, indicating a possible inequity in care. To address this, we implemented a comprehensive in-EHR tracking system to follow patients along their diagnostic testing odyssey. Using EHR access log data, we found use of the system significantly speeded review of results and significantly increased documentation of follow-up recommendations.
With the onset of the COVID-19 pandemic, pediatric genetics evaluation quickly transitioned to telemedicine. We worried that subtle dysmorphology physical examination information lost over video might decrease diagnostic effectiveness. We performed a large analysis of telemedicine practice and found that patients seen by telemedicine were more likely to report non-Hispanic white background and come from wealthier neighborhoods. Thus, there may be inequities in access to telemedicine-based genetics care, similar to findings in other fields. We did not, however, identify a significant difference in clinical effectiveness from a new molecular diagnosis perspective.
Campbell IM, McManus ML, Cusick FC, Junod DC, Sheppard SE, Lourie EM, Shelov MD, Luberti AA. Clinical Decision Support with a Comprehensive In-EHR Patient Tracking System to Improve Inpatient Genetic Testing Follow Up. Presented at the American Medical Informatics Association 2021 Annual Symposium, San Diego, CA.
Campbell IM, Crowley TB, Keena B, Donoghue S, McManus ML, Zackai EH. The Experience of One Pediatric Geneticist with Telemedicine-Based Clinical Diagnosis. Am J Med Genet A. 2022 Jul 30. Online ahead of print. PMID: 35906847
Szigety KM, et al. Clinical Effectiveness of Telemedicine-Based Pediatric Genetics Care. Pediatrics. 2022 Jun 1;e2021054520. PMID: 35642503
Children’s Hospital of Philadelphia recently implemented discrete genetic variant capture, genetic problem annotation, and clinical decision support based on genetic conditions in our EHR. This helps clinicians to improve the care of their patients with genetic disease without explicit prior knowledge of specific recommendations. Developing safe clinical decision support interventions that have high clinical impact on patient care relies on understanding the human factors within the complex sociotechnical system. A major goal of the lab is the ongoing deployment of new content for more diseases, identification of historically identified patients who can benefit from these interventions, and ongoing maintenance and review of existing content.